13 Who’s that pathogen?!
⬜ Developing Hypotheses
⬜ Sample Collection
⬜ Outbreak Investigation
🟩 Sequencing
⬜ Bioinformatics
⬜ Molecular Epidemiology
⬜ Public Health Implementation
After much discussion, the multidisciplinary team decided that since the bat trainers were the earliest known cases to exhibit flu-like symptoms but test negative for flu, these samples would be ideal to start with. The multidisciplinary team discussed amongst themselves which kinds of sequencing approaches would be best to help identify the presence of microorganisms in the samples. Since the initial testing and sequencing that was performed on the muubats did not detect any pathogens known to cause any human illnesses, the team thought it would be best to start broad. The Bioinformaticians and Microbiologists decided to take a shotgun metagenomics approach and sequence everything in the samples collected from the bat trainers.
Shotgun metagenomics sequencing allows microbiologists to sample all organisms in a mixed and complex sample. This sequencing and bioinformatics approach is comprehensive and allows us to evaluate microbial diversity, that is identify the types of species in a sample, as well as detect the relative abundance of certain microbes in the sample. Relative abundance is an inferred estimate of the percentage composition of a particular kind of organism relative to the totality of organisms detected and identified in a mixed sample. Oftentimes relative abundances measure on a scale from 0 to 1.
The Microbiologists sequenced everything that could be present in the samples using shotgun metagenomics. Using this approach, nucleic acids, such as DNA and RNA is fragmented into smaller components which are then independently sequenced. Once the sequencing is complete the Bioinformaticians performed QC on the raw data to check for contaminants and background noise. Once everything looked good, the Bioinformaticians proceeded to process the raw sequencing data using their metagenomic pipelines. Their pipelines have been rigorously validated and was specially configured with tools to identify microbial species against the Internal Sequencing Database that is maintained by the Regional Health Lab, as well as a built-in algorithm that infers the relative abundance of microorganisms in the sample.
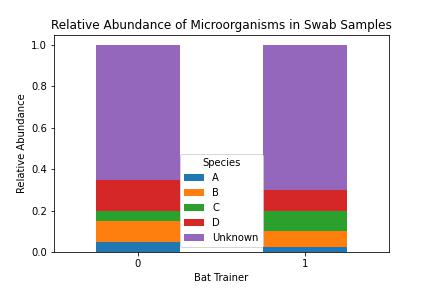
Relative Abundance of Microorganisms in Swab Samples of Bat Trainers Collected 31-01-08. The relative abundance chart shows the percent composition in terms of the proportions of species in the swab samples taken from the Bat Trainers. The relative abundance is plotted on the vertical axis, with the Bat Trainers on the horizontal axis. Species were identified using the Internal Sequencing Database maintained by the Regional Health Lab.
13.1 Discussion Six: Interpretation Metagenomics Sequencing Results and Visualizations
The figure above shows the relative abundance of microbial species detected in swab samples from the bat trainers, based on metagenomic sequencing data.
- How many dominant species can you identify, and what might their relative abundance suggest about the microbial community composition?
- Were any reads unassigned or classified as “unknown” because they did not match any organisms in the Internal Sequencing Database?
- If so, what are some possible explanations for these unclassified sequences (e.g., novel organisms, incomplete reference data, contamination)?
- How would you distinguish between a truly novel pathogen and background microbial noise in a complex sample like this?
- What additional analyses or data would help you validate your interpretation?
Metagenomic data often contain a mix of informative signals and environmental noise. The challenge is not just identifying what’s present, but deciding what’s relevant.