Code
terminal
git clone https://github.com/NW-PaGe/private-lineages.gitBelow there are two ways to install this software, either on your local machine (see installation) or via a Github Codespace which does not require any software to be installed on your personal machine (see Github Codespace)
lineages_test.Rproj R project:Can double click on the file or open in a terminal window by typing
renvIn the Rstudio window’s console, execute
If you want to run the code but don’t want to install anything on your local machine, use a Github Codespace.
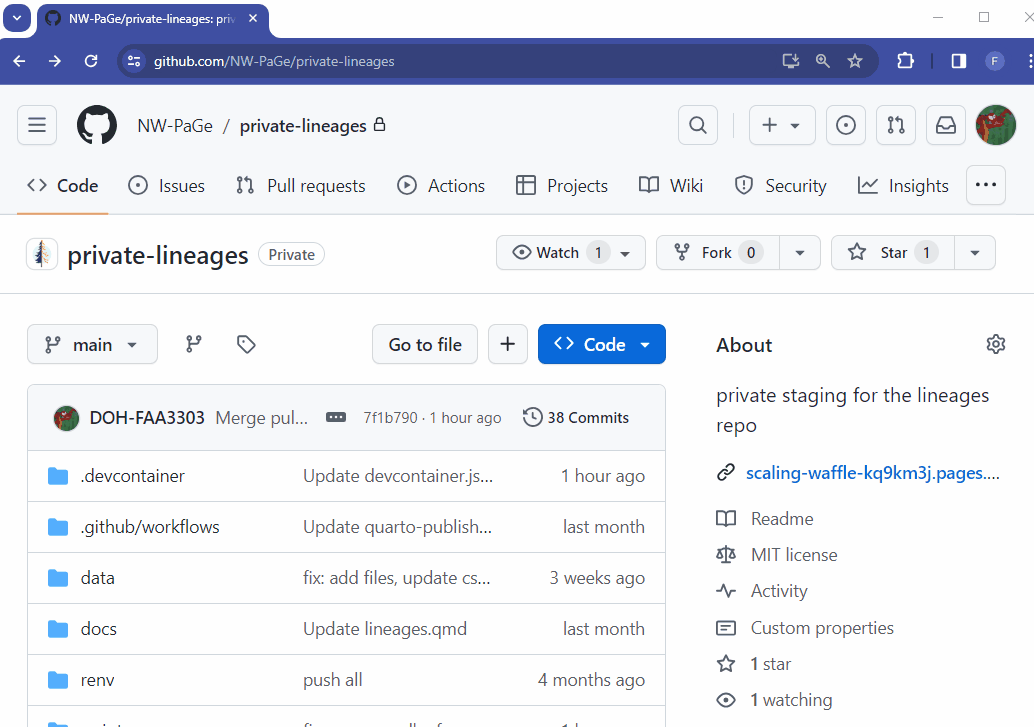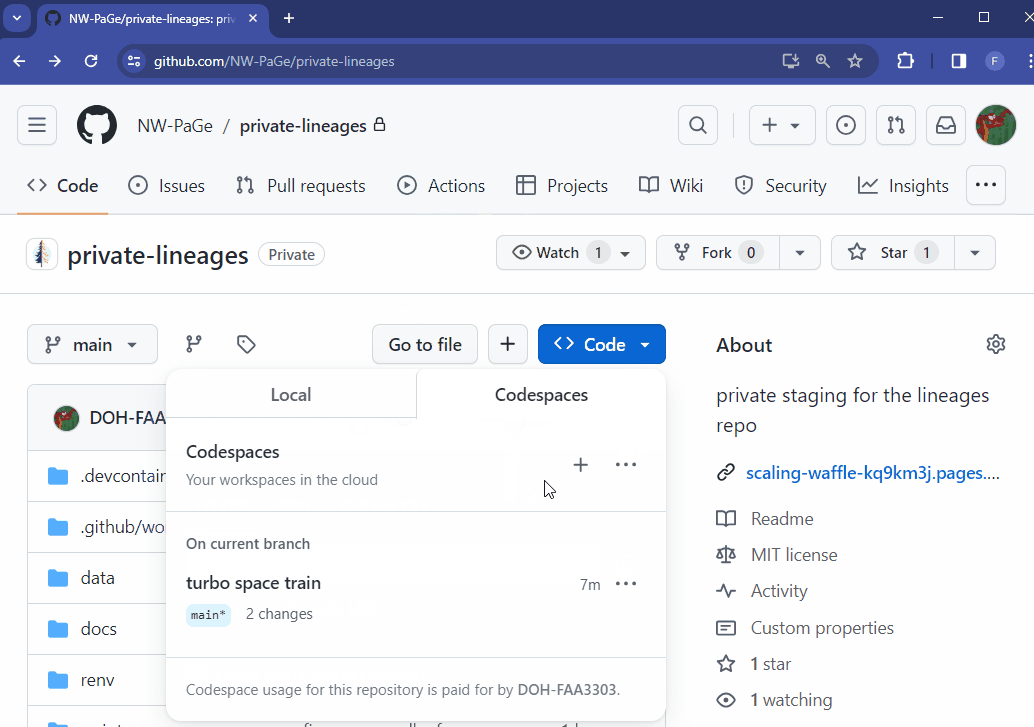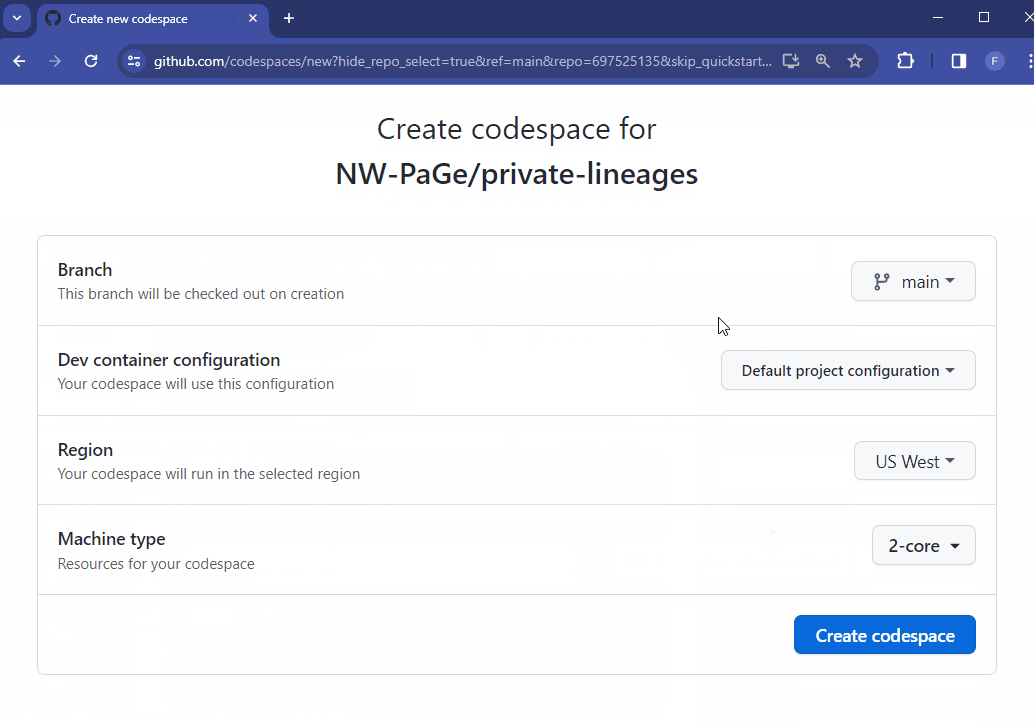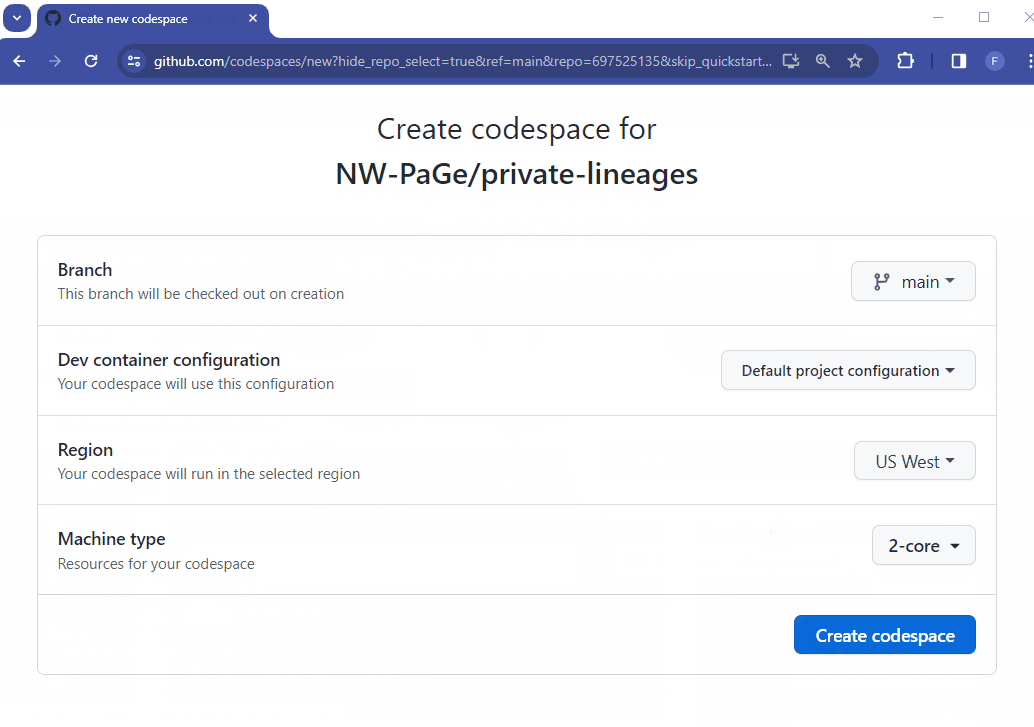
renv::restore() and follow its instructions. It should download all the packages in the renv folder in this repoNow you can run this code in a browser window without needing to install any software on your computer 😎
The repo will output a .csv file called lineages.csv that will be updated whenever new lineages are updated from the CDC. You can refresh/fetch this git repo for new changes and the .csv file will be updated
To run the process:
The variables produced by the scripts are used in the Sequencing and Variants Report. Here’s a list:
| Variables | Description |
|---|---|
| cdc_class | Variable indicating VOC (variant of concern) or VBM (variant being monitored) |
| who_name | Variable indicating the WHO name |
| doh_variant_name | Grouping variable in WA DOH Sequencing and Variant report |
| hex_code | Hex color for doh_variant_name group |
| lineage_reporting_group | Variable indicating reporting group of lineage coded as: 1: Currently monitoring 2: Formerly monitoring 3: Formerly circulating, not monitored |
| report_table_name | Variable name in numerical/pango form for table outputs |
For details on how to contribute to this project, please read the NW-PaGe contributing guide
This repo uses an MIT license